parallel_load_dataframes_s3
To get started:
Dynamically pull and run
from hamilton import dataflows, driver
from hamilton.execution import executors
# downloads into ~/.hamilton/dataflows and loads the module -- WARNING: ensure you know what code you're importing!
parallel_load_dataframes_s3 = dataflows.import_module("parallel_load_dataframes_s3", "elijahbenizzy")
# Switch this out for ray, dask, etc. See docs for more info.
remote_executor = executors.MultiThreadingExecutor(max_tasks=20)
dr = (
driver.Builder()
.enable_dynamic_execution(allow_experimental_mode=True)
.with_remote_executor(remote_executor)
.with_config({}) # replace with configuration as appropriate
.with_modules(parallel_load_dataframes_s3)
.build()
)
# If you have sf-hamilton[visualization] installed, you can see the dataflow graph
# In a notebook this will show an image, else pass in arguments to save to a file
# dr.display_all_functions()
# Execute the dataflow, specifying what you want back. Will return a dictionary.
result = dr.execute(
[parallel_load_dataframes_s3.CHANGE_ME, ...], # this specifies what you want back
inputs={...} # pass in inputs as appropriate
)
Use published library version
pip install sf-hamilton-contrib --upgrade # make sure you have the latest
from hamilton import dataflows, driver
from hamilton.execution import executors
# Make sure you've done - `pip install sf-hamilton-contrib --upgrade`
from hamilton.contrib.user.elijahbenizzy import parallel_load_dataframes_s3
# Switch this out for ray, dask, etc. See docs for more info.
remote_executor = executors.MultiThreadingExecutor(max_tasks=20)
dr = (
driver.Builder()
.enable_dynamic_execution(allow_experimental_mode=True)
.with_remote_executor(remote_executor)
.with_config({}) # replace with configuration as appropriate
.with_modules(parallel_load_dataframes_s3)
.build()
)
# If you have sf-hamilton[visualization] installed, you can see the dataflow graph
# In a notebook this will show an image, else pass in arguments to save to a file
# dr.display_all_functions()
# Execute the dataflow, specifying what you want back. Will return a dictionary.
result = dr.execute(
[parallel_load_dataframes_s3.CHANGE_ME, ...], # this specifies what you want back
inputs={...} # pass in inputs as appropriate
)
Modify for your needs
Now if you want to modify the dataflow, you can copy it to a new folder (renaming is possible), and modify it there.
dataflows.copy(parallel_load_dataframes_s3, "path/to/save/to")
Purpose of this module
This module loads up dataframes specified in a json or jsonl set of files from s3.
You have to pass it:
- The bucket to download from (
bucket) - The path within the bucket to crawl (
path_in_bucket) - The caching directory for saving files locally (
save_dir)
Optionally, you can pass it:
- The AWS profile (
aws_profile), this defaults todefault
This will look for all files that end with json or jsonl, under s3://<bucket>/<path_within_bucket> download them to the save_dir, load them, and concatenate it into a dataframe.
Note that if a file exists locally it is skipped -- thus this is idempotent.
Configuration Options
N/A
Limitations
This only downloads json/jsonl files, and currently crawls the entire sub-bucket. This also requires
that all files be able to be held in memory, and are of a uniform schema.
Source code
__init__.py
import dataclasses
import logging
import os
from pathlib import Path
from typing import List
logger = logging.getLogger(__name__)
from hamilton import log_setup
log_setup.setup_logging(logging.INFO)
from hamilton import contrib
with contrib.catch_import_errors(__name__, __file__, logger):
# non-hamilton imports go here
import boto3
import pandas as pd
from boto3 import Session
pass
# hamilton imports go here; check for required version if need be.
from hamilton.htypes import Collect, Parallelizable
# from hamilton.log_setup import setup_logging
logger = logging.getLogger(__name__)
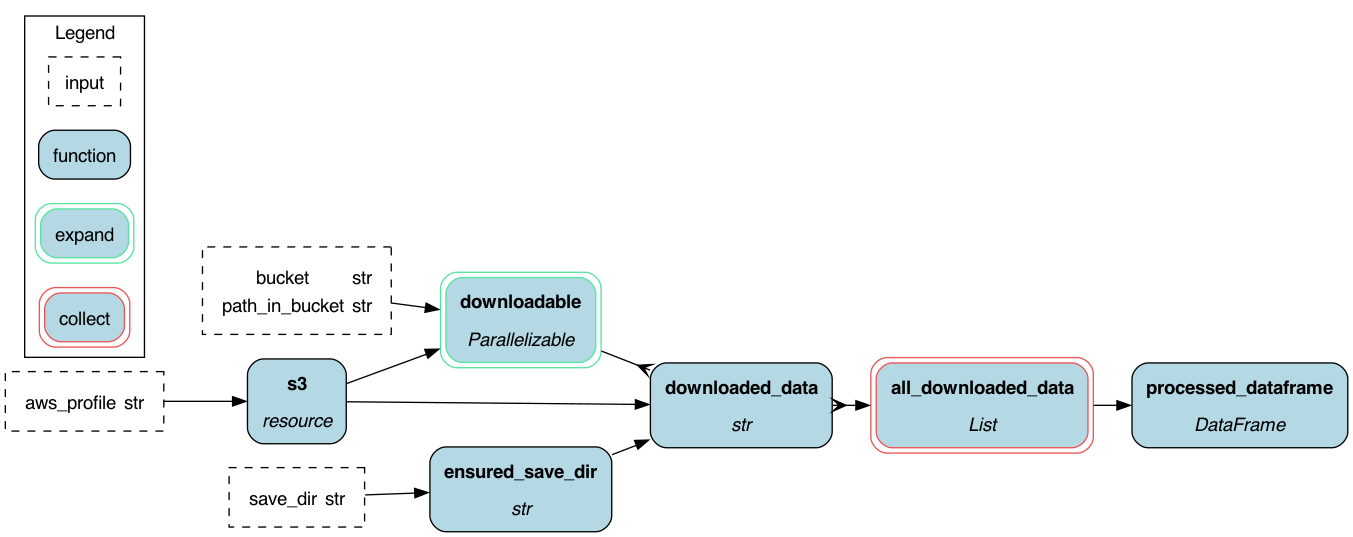
def s3(aws_profile: str = "default") -> boto3.resource:
"""Returns a boto3 resource for the 'aws_profile' profile"""
session = Session(profile_name=aws_profile)
return session.resource("s3")
@dataclasses.dataclass
class ToDownload:
"""Simple dataclass to contain downloading files"""
key: str
bucket: str
def ensured_save_dir(save_dir: str) -> str:
"""Ensures that a savedir exists for later"""
if not os.path.exists(save_dir):
Path(save_dir).mkdir()
return save_dir
def downloadable(
s3: boto3.resource, bucket: str, path_in_bucket: str
) -> Parallelizable[ToDownload]:
"""Lists downloadables from the s3 bucket"""
bucket_obj = s3.Bucket(bucket)
objs = list(bucket_obj.objects.filter(Prefix=path_in_bucket).all())
objs = [obj for obj in objs if obj.key.endswith(".jsonl") or obj.key.endswith(".json")]
logger.info(f"Found {len(objs)} objects in {bucket}/{path_in_bucket}")
for obj in objs:
yield ToDownload(key=obj.key, bucket=bucket)
def _already_downloaded(path: str) -> bool:
"""Checks if the data is already downloaded"""
if os.path.exists(path):
return True
return False
def downloaded_data(
downloadable: ToDownload,
ensured_save_dir: str,
s3: boto3.resource,
) -> str:
"""Downloads data, short-circuiting if the data already exists locally"""
download_location = os.path.join(ensured_save_dir, downloadable.key)
if _already_downloaded(download_location):
logger.info(f"Already downloaded {download_location}")
return download_location
parent_path = os.path.dirname(download_location)
if not os.path.exists(parent_path):
os.makedirs(parent_path, exist_ok=True)
# This works with threading, but might not work in parallel with multiprocessing
# TODO -- use a connection pool
bucket = s3.Bucket(downloadable.bucket)
bucket.download_file(downloadable.key, download_location)
logger.info(f"Downloaded {download_location}")
return download_location
def all_downloaded_data(downloaded_data: Collect[str]) -> List[str]:
"""Returns a list of all downloaded locations"""
out = []
for path in downloaded_data:
out.append(path)
return out
def _jsonl_parse(path: str) -> pd.DataFrame:
"""Loads a jsonl file into a dataframe"""
return pd.read_json(path, lines=True)
def processed_dataframe(all_downloaded_data: List[str]) -> pd.DataFrame:
"""Processes everything into a dataframe"""
out = []
for floc in all_downloaded_data:
out.append(_jsonl_parse(floc))
return pd.concat(out)
if __name__ == "__main__":
# Code to create an imaging showing on DAG workflow.
# run as a script to test Hamilton's execution
import __init__ as parallel_load_dataframes_s3
from hamilton import driver
dr = (
driver.Builder()
.with_modules(parallel_load_dataframes_s3)
.enable_dynamic_execution(allow_experimental_mode=True)
.build()
)
# saves to current working directory creating dag.png.
dr.display_all_functions("dag", {"format": "png", "view": False})
Requirements
# put non-hamilton requirements here
boto3
pandas