customize_embeddings
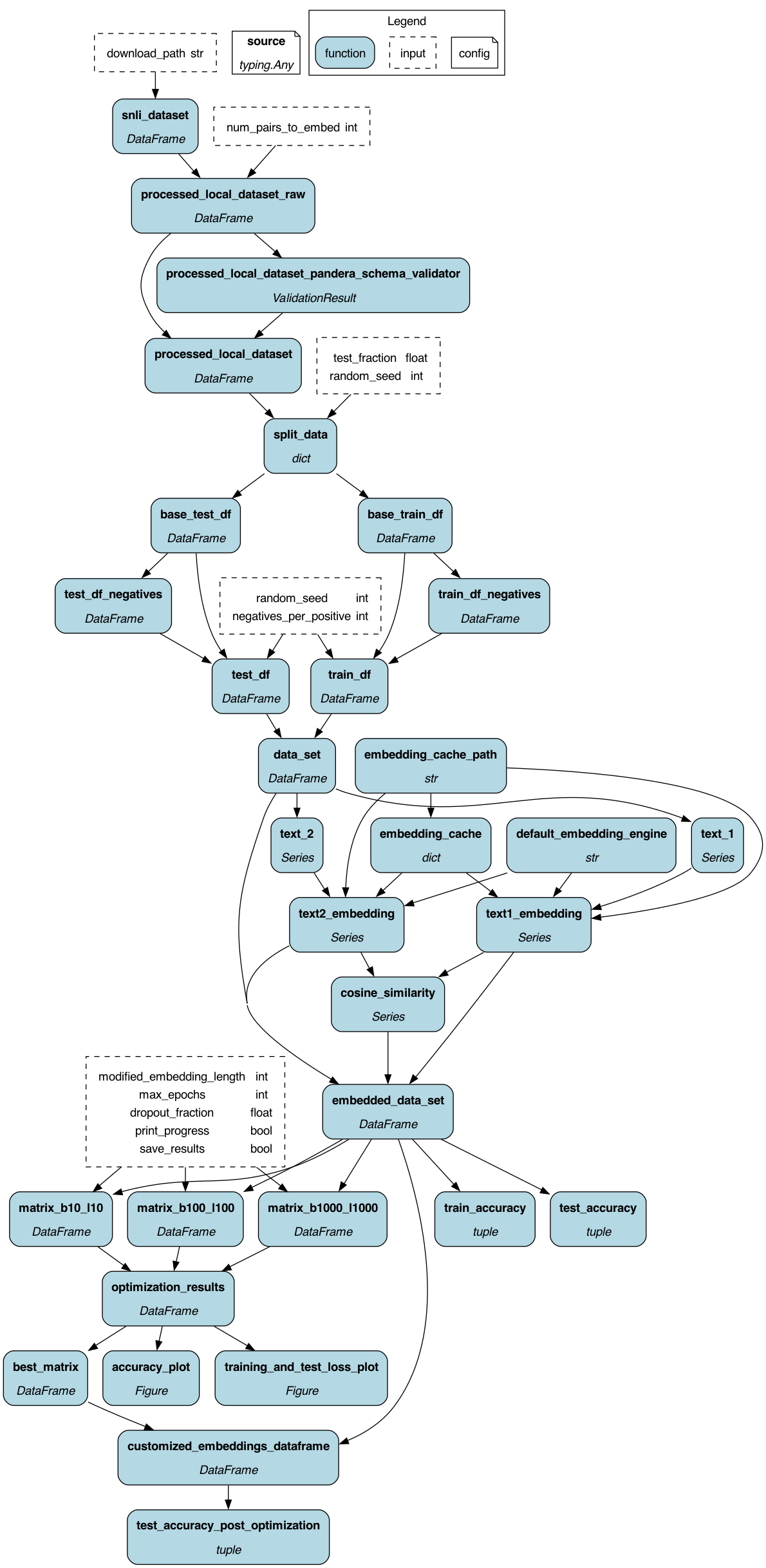
To get started:
Dynamically pull and run
from hamilton import dataflows, driver
# downloads into ~/.hamilton/dataflows and loads the module -- WARNING: ensure you know what code you're importing!
customize_embeddings = dataflows.import_module("customize_embeddings", "skrawcz")
dr = (
driver.Builder()
.with_config({}) # replace with configuration as appropriate
.with_modules(customize_embeddings)
.build()
)
# If you have sf-hamilton[visualization] installed, you can see the dataflow graph
# In a notebook this will show an image, else pass in arguments to save to a file
# dr.display_all_functions()
# Execute the dataflow, specifying what you want back. Will return a dictionary.
result = dr.execute(
[customize_embeddings.CHANGE_ME, ...], # this specifies what you want back
inputs={...} # pass in inputs as appropriate
)
Use published library version
pip install sf-hamilton-contrib --upgrade # make sure you have the latest
from hamilton import dataflows, driver
# Make sure you've done - `pip install sf-hamilton-contrib --upgrade`
from hamilton.contrib.user.skrawcz import customize_embeddings
dr = (
driver.Builder()
.with_config({}) # replace with configuration as appropriate
.with_modules(customize_embeddings)
.build()
)
# If you have sf-hamilton[visualization] installed, you can see the dataflow graph
# In a notebook this will show an image, else pass in arguments to save to a file
# dr.display_all_functions()
# Execute the dataflow, specifying what you want back. Will return a dictionary.
result = dr.execute(
[customize_embeddings.CHANGE_ME, ...], # this specifies what you want back
inputs={...} # pass in inputs as appropriate
)
Modify for your needs
Now if you want to modify the dataflow, you can copy it to a new folder (renaming is possible), and modify it there.
dataflows.copy(customize_embeddings, "path/to/save/to")
Purpose of this module
This module is used to customize embeddings for text data. It is based on MIT licensed code from this OpenAI cookbook.
The output is a matrix that you can use to multiply your embeddings. The product of this multiplication is a 'custom embedding' that will better emphasize aspects of the text relevant to your use case. In binary classification use cases, the OpenAI cook book author claims to have seen error rates drop by as much as 50%.
Why customize embeddings? Embeddings are a way to represent text as a vector of numbers. The numbers are chosen so that similar words have similar vectors. For example, the vectors for "cat" and "dog" will be closer together than the vectors for "cat" and "banana". Embeddings are useful for many NLP tasks, such as sentiment analysis, text classification, and question answering. Embeddings are often trained on a large corpus of text, such as Wikipedia. However, these embeddings may not be optimal for a specific task. E.g. you may want to be able to distinguish between "cat" and "dog" for a specific task, but the embeddings may not allow you to do that reliably. This module allows you to customize some embeddings for a specific task, e.g. by making the vectors for "cat" and "dog" further apart.
How might I use this module?
If you pass in {"source":"snli"} as configuration to the driver, the module will download the corpus and unzip it and
use that as input to optimize embeddings for. The embeddings of sentences are optimized for the task of predicting
whether a sentence pair is "entailment", or "contradiction". That is, each pair of sentences are logically entailed
(i.e., one implies the other). These pairs are our positives (label = 1). We generate synthetic negatives by combining
sentences from different pairs, which are presumed to not be logically entailed (label = -1).
If you pass in {"source":"local"} as configuration to the driver, the module will load a local dataset you provide a
path to. The dataset should be a csv with columns "text_1", "text_2", and "label". The label should be +1 if the text
pairs are similar and -1 if the text pairs are dissimilar.
Otherwise if you pass in {} as configuration to the driver, the module will require you to pass in a dataframe as
processed_local_dataset as an input. The dataframe should have columns "text_1", "text_2", and "label". The label should be +1 if the
text pairs are similar and -1 if the text pairs are dissimilar.
In general to use this module you'll need to do the following:
- Create a dataset of text pairs, where each pair is either similar or dissimilar. Mechanically that means you'll need
to modify/override
processed_local_dataset()so that you output a dataset of [text_1, text_2, label] where label is +1 if the pairs of text are similar and -1 if the pairs of text are dissimilar. See the docstring forprocessed_local_dataset(). - Modify the "hyperparameters" for a couple of the functions, e.g.
optimize_matrix(), to suit your needs. - Modify how embeddings are generated, e.g.
_get_embedding(), to suit your needs. - Adjust any logic for creating negative pairs, that is, modify
_generate_negative_pairs(), to suit your needs. For example, if you have multi-class labels you'll have to modify how you generate negative pairs. - Profit!
To execute the DAG, the recommended outputs to grab are:
outputs=[
"train_accuracy",
"test_accuracy",
"embedded_dataset_histogram",
"test_accuracy_post_optimization",
"accuracy_plot",
"training_and_test_loss_plot",
"customized_embeddings_dataframe",
"customized_dataset_histogram",
]
You should be able to read the module top to bottom which corresponds roughly to the order of execution.
What type of functionality is in this module?
The module includes functions for getting embeddings, calculating cosine similarity, processing datasets, splitting data,
generating negative pairs, optimizing matrices, and plotting results. It also shows how one can use @config.when
to swap out how the data is loaded, @check_output for schema validation of a dataframe, @parameterize for
creating many functions from a single function, and @inject to collect results from the results of @parameterize.
The module uses libraries such as numpy, pandas, plotly, torch, sklearn, and openai.
Configuration Options
{"source": "snli"} will use the Stanford SNLI corpus. Use this to run the code as-is.
{"source": "local"} will load a local dataset you provide a path to.
{} will require you to pass in a dataframe with name raw_data as an input.
Limitations
This code is currently set up to work with OpenAI. It could be modified to work with other embedding producers/providers.
The matrix optimization is not set up to be parallelized, though it could be done using Hamilton constructs,
like Parallelizeable.
Source code
__init__.py
"""
The code in this module is based on MIT licensed code from the OpenAI cookbook. Since
this module uses a substantial portion of it, we are including the copyright notice below
as required.
----------------------------------------------------------------------------------------------
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
----------------------------------------------------------------------------------------------
"""
import logging
import os
import pickle # for saving the embeddings cache
import random # for generating run IDs
import sys
import zipfile
from typing import List, Tuple # for type hints
logger = logging.getLogger(__name__)
from hamilton import contrib
with contrib.catch_import_errors(__name__, __file__, logger):
import numpy as np # for manipulating arrays
import openai
import pandas as pd # for manipulating data in dataframes
import pandera as pa # for data validation
import plotly.express as px # for plots
import plotly.graph_objs as go # for plot object type
import requests
import torch # for matrix optimization
from sklearn.model_selection import (
train_test_split,
) # for splitting train & test data
from tenacity import retry, stop_after_attempt, wait_random_exponential
from hamilton.function_modifiers import (
check_output,
config,
extract_columns,
extract_fields,
group,
inject,
load_from,
parameterize,
source,
value,
)
client = openai.OpenAI()
@retry(wait=wait_random_exponential(min=1, max=20), stop=stop_after_attempt(6))
def _get_embedding(text: str, model="text-similarity-davinci-001", **kwargs) -> List[float]:
"""Get embedding from OpenAI API.
:param text: text to embed.
:param model: the embedding model to use.
:param kwargs: any additional kwargs to pass to the API.
:return: list of floats representing the embedding.
"""
# replace newlines, which can negatively affect performance.
text = text.replace("\n", " ")
response = client.embeddings.create(input=[text], model=model, **kwargs)
return response.data[0].embedding
def _cosine_similarity(a, b):
"""Compute cosine similarity between two vectors."""
return np.dot(a, b) / (np.linalg.norm(a) * np.linalg.norm(b))
def embedding_cache_path() -> str:
"""Path to embeddings cache. Use `overrides={"embedding_cache_path": "my_path"}` to override."""
return "data/snli_embedding_cache.pkl" # embeddings will be saved/loaded here
def default_embedding_engine() -> str:
"""Default embedding engine. Use `overrides={"default_embedding_engine": "my_engine"}` to override."""
return "babbage-similarity" # text-embedding-ada-002 is recommended
@config.when(source="snli")
def snli_dataset(download_path: str = "data") -> pd.DataFrame:
"""Uses the SNLI dataset, downloading it and unzipping to data/, if necessary."""
if os.path.exists(f"{download_path}/snli_1.0/snli_1.0_train.txt"):
logger.info("Not downloading - snli data already exists.")
return pd.read_csv(f"{download_path}/snli_1.0/snli_1.0_train.txt", delimiter="\t")
# URL of the file to be downloaded
url = "https://nlp.stanford.edu/projects/snli/snli_1.0.zip"
# Send a HTTP request to the URL of the file, stream = True means download the file
response = requests.get(url, stream=True)
# File to save the downloaded file
file_name = "snli_1.0.zip"
# Write the downloaded file into "snli_1.0.zip"
with open(file_name, "wb") as fd:
for chunk in response.iter_content(chunk_size=1024):
fd.write(chunk)
# Create a ZipFile Object
with zipfile.ZipFile(file_name) as zip_file:
# Extract all the contents of zip file in current directory
zip_file.extractall(path="data")
# Remove the downloaded zip file
os.remove(file_name)
logger.info(f"Downloaded SNLI data and saved to {os.getcwd()}/data/")
return pd.read_csv(f"{download_path}/snli_1.0/snli_1.0_train.txt", delimiter="\t")
# you can add a schema to validate your data
processed_dataset_schema = pa.DataFrameSchema(
{
"text_1": pa.Column(str, nullable=False),
"text_2": pa.Column(str, nullable=False),
"label": pa.Column(int, nullable=False, checks=[pa.Check.isin([-1, 1])]),
},
strict=True,
)
@config.when(source="snli")
@check_output(schema=processed_dataset_schema, importance="fail")
def processed_local_dataset__snli(
snli_dataset: pd.DataFrame,
num_pairs_to_embed: int = 1000, # 1000 is arbitrary
) -> pd.DataFrame:
"""Processes a raw dataset into a dataframe of text pairs to embed; and check that it matches the schema.
This assumes the SNLI dataset, which has 3 columns: sentence1, sentence2, gold_label.
Override this with a dataframe (`overrides={"processed_local_dataset": my_df}`) if you have your own dataset
that's ready, or modify it to match your use case.
:param snli_dataset: the snli dataset to process.
:param num_pairs_to_embed: the number of pairs to embed. Default 1000. 1000 is arbitrary.
:return: dataframe of text pairs with labels. Schema: text_1, text_2, label (1 for similar, -1 for dissimilar)
"""
# you can customize this to preprocess your own dataset
# output should be a dataframe with 3 columns: text_1, text_2, label (1 for similar, -1 for dissimilar)
df = snli_dataset[snli_dataset["gold_label"].isin(["entailment"])]
df["label"] = df["gold_label"].apply(lambda x: {"entailment": 1, "contradiction": -1}[x])
df = df.rename(columns={"sentence1": "text_1", "sentence2": "text_2"})
df = df[["text_1", "text_2", "label"]]
df = df.head(num_pairs_to_embed)
return df
@config.when(source="local")
@check_output(schema=processed_dataset_schema, importance="fail")
@load_from.csv(
path=source("local_dataset_path")
# see data loader docuemntation and the PandasCSVReader for values you can pass in:
# - https://hamilton.dagworks.io/en/latest/reference/io/available-data-adapters/#data-loaders
# - https://github.com/dagworks-inc/hamilton/blob/main/hamilton/plugins/pandas_extensions.py#L89-L255
)
def processed_local_dataset__local(
local_dataset: pd.DataFrame,
) -> pd.DataFrame:
"""Uses a local dataset, reading it from the given path.
Override this with a dataframe (`overrides={"processed_local_dataset": my_df}`) if you have your own dataset
that's ready in a notebook, or modify it to match your use case.
:param local_dataset: dataframe loaded by the Pandas CSV Reader.
:return: dataframe of text pairs with labels. Schema: text_1, text_2, label (1 for similar, -1 for dissimilar).
"""
return local_dataset
# split data into train and test sets
@extract_fields({"base_train_df": pd.DataFrame, "base_test_df": pd.DataFrame})
def split_data(
processed_local_dataset: pd.DataFrame,
test_fraction: float = 0.5, # 0.5 is fairly arbitrary
random_seed: int = 123, # random seed is arbitrary, but is helpful in reproducibility
) -> dict:
"""Splits a processed dataset into train and test sets.
Note that it's important to split data into training and test sets before generating synthetic negatives or
positives. You don't want any text strings in the training data to show up in the test data.
If there's contamination, the test metrics will look better than they'll actually be in production.
"""
train_df, test_df = train_test_split(
processed_local_dataset,
test_size=test_fraction,
stratify=processed_local_dataset["label"],
random_state=random_seed,
)
train_df.loc[:, "dataset"] = "train"
test_df.loc[:, "dataset"] = "test"
return {"base_train_df": train_df, "base_test_df": test_df}
def _dataframe_of_negatives(dataframe_of_positives: pd.DataFrame) -> pd.DataFrame:
"""Return dataframe of negative pairs made by combining elements of positive pairs.
This is another piece of the code that you will need to modify to match your use case.
If you have data with positives and negatives, you can skip this section.
If you have data with only positives, you can mostly keep it as is, where it generates negatives only.
If you have multiclass data, you will want to generate both positives and negatives.
The positives can be pairs of text that share labels, and the negatives can be pairs of text that
do not share labels.
The final output should be a dataframe with text pairs, where each pair is labeled -1 or 1.
"""
texts = set(dataframe_of_positives["text_1"].values) | set(
dataframe_of_positives["text_2"].values
)
all_pairs = {(t1, t2) for t1 in texts for t2 in texts if t1 < t2}
positive_pairs = set(
tuple(text_pair) for text_pair in dataframe_of_positives[["text_1", "text_2"]].values
)
negative_pairs = all_pairs - positive_pairs
df_of_negatives = pd.DataFrame(list(negative_pairs), columns=["text_1", "text_2"])
df_of_negatives["label"] = -1
return df_of_negatives
def train_df_negatives(base_train_df: pd.DataFrame) -> pd.DataFrame:
"""Return dataframe of negative pairs for training."""
_df = _dataframe_of_negatives(base_train_df)
_df["dataset"] = "train"
return _df
def test_df_negatives(base_test_df: pd.DataFrame) -> pd.DataFrame:
"""Return dataframe of negative pairs for testing."""
_df = _dataframe_of_negatives(base_test_df)
_df["dataset"] = "test"
return _df
@parameterize(
train_df={
"base_df": source("base_train_df"),
"df_negatives": source("train_df_negatives"),
},
test_df={
"base_df": source("base_test_df"),
"df_negatives": source("test_df_negatives"),
},
)
def construct_df(
base_df: pd.DataFrame,
df_negatives: pd.DataFrame,
negatives_per_positive: int = 1,
random_seed: int = 123,
) -> pd.DataFrame:
"""Return dataframe of {base_df} paris with negatives added."""
return pd.concat(
[
base_df,
df_negatives.sample(n=len(base_df) * negatives_per_positive, random_state=random_seed),
]
)
# def test_df(
# base_test_df: pd.DataFrame,
# test_df_negatives: pd.DataFrame,
# negatives_per_positive: int = 1,
# random_seed: int = 123,
# ) -> pd.DataFrame:
# """Return dataframe of testing pairs, with negatives added."""
# return pd.concat(
# [
# base_test_df,
# test_df_negatives.sample(
# n=len(base_test_df) * negatives_per_positive, random_state=random_seed
# ),
# ]
# )
# Expose text_1 and text_2 columns from train and test dataframes
@extract_columns("text_1", "text_2")
def data_set(train_df: pd.DataFrame, test_df: pd.DataFrame) -> pd.DataFrame:
"""Combine train and test dataframes into one. Rather than passing around two dataframes,
we just choose to have one that we subset as needed into train or test.
"""
_df = pd.concat([train_df, test_df])
_df.reset_index(inplace=True) # reset index to avoid duplicate indices
return _df
# this function will get embeddings from the cache and save them there afterward
def _get_embedding_with_cache(
text: str,
engine: str,
embedding_cache: dict = None,
embedding_cache_path: str = None,
) -> list:
"""Get embedding from cache if available, otherwise call API to get embedding and save to cache."""
if embedding_cache is None:
embedding_cache = {}
if (text, engine) not in embedding_cache.keys():
# if not in cache, call API to get embedding
embedding_cache[(text, engine)] = _get_embedding(text, engine)
# save embeddings cache to disk after each update
with open(embedding_cache_path, "wb") as embedding_cache_file:
pickle.dump(embedding_cache, embedding_cache_file)
return embedding_cache[(text, engine)]
def embedding_cache(embedding_cache_path: str) -> dict:
"""Establish a cache of embeddings to avoid recomputing
Cache is a dict of tuples (text, engine) -> embedding
:param embedding_cache_path: path to save/load cache
:return: cache of embeddings.
"""
try:
with open(embedding_cache_path, "rb") as f:
embedding_cache = pickle.load(f)
except FileNotFoundError:
precomputed_embedding_cache_path = (
"https://cdn.openai.com/API/examples/data/snli_embedding_cache.pkl"
)
embedding_cache = pd.read_pickle(precomputed_embedding_cache_path)
return embedding_cache
def text1_embedding(
text_1: pd.Series,
embedding_cache_path: str,
embedding_cache: dict,
default_embedding_engine: str,
) -> pd.Series:
"""Get embeddings for text_1 column of a dataframe."""
_col = text_1.apply(
_get_embedding_with_cache,
engine=default_embedding_engine,
embedding_cache_path=embedding_cache_path,
embedding_cache=embedding_cache,
)
_col.name = "text_1_embedding"
return _col
def text2_embedding(
text_2: pd.Series,
embedding_cache_path: str,
embedding_cache: dict,
default_embedding_engine: str,
) -> pd.Series:
"""Get embeddings for text_2 column of a dataframe."""
_col = text_2.apply(
_get_embedding_with_cache,
engine=default_embedding_engine,
embedding_cache_path=embedding_cache_path,
embedding_cache=embedding_cache,
)
_col.name = "text_2_embedding"
return _col
def cosine_similarity(text1_embedding: pd.Series, text2_embedding: pd.Series) -> pd.Series:
"""Computed cosine similarity between text1_embedding and text2_embedding.
Here we measure similarity of text using cosine similarity. In our experience, most distance functions
(L1, L2, cosine similarity) all work about the same. Note if your embeddings are already normalized to length 1,
the cosine similarity is equivalent to dot product.
"""
similarity_scores = text1_embedding.combine(
text2_embedding, lambda x1, x2: 1 - _cosine_similarity(x1, x2)
)
similarity_scores.name = "cosine_similarity"
return similarity_scores
def embedded_data_set(
data_set: pd.DataFrame,
text1_embedding: pd.Series,
text2_embedding: pd.Series,
cosine_similarity: pd.Series,
) -> pd.DataFrame:
"""Combined DF of data_set, text1_embedding, text2_embedding, and cosine_similarity."""
_df = pd.concat([data_set, text1_embedding, text2_embedding, cosine_similarity], axis=1)
return _df
def _accuracy_and_se(
cosine_similarity: list[float], labeled_similarity: list[int]
) -> Tuple[float, float]:
"""Calculate accuracy (and its standard error) of predicting label=1 if similarity>x
x is optimized by sweeping from -1 to 1 in steps of 0.01
:param cosine_similarity: list of cosine similarity scores
:param labeled_similarity: list of labels (1 or -1)
:return: tuple of accuracy and standard error
"""
accuracies = []
for threshold_thousandths in range(-1000, 1000, 1):
threshold = threshold_thousandths / 1000
total = 0
correct = 0
for cs, ls in zip(cosine_similarity, labeled_similarity):
total += 1
if cs > threshold:
prediction = 1
else:
prediction = -1
if prediction == ls:
correct += 1
accuracy = correct / total
accuracies.append(accuracy)
a = max(accuracies)
n = len(cosine_similarity)
standard_error = (a * (1 - a) / n) ** 0.5 # standard error of binomial
return a, standard_error
def embedded_dataset_histogram(embedded_data_set: pd.DataFrame) -> go.Figure:
"""Plot histogram of cosine similarities.
The graphs show how much the overlap there is between the distribution of cosine similarities for similar and
dissimilar pairs. If there is a high amount of overlap, that means there are some dissimilar pairs with greater
cosine similarity than some similar pairs.
"""
_plot = px.histogram(
embedded_data_set,
x="cosine_similarity",
color="label",
barmode="overlay",
width=500,
facet_row="dataset",
)
return _plot
@parameterize(
**{
"train_accuracy": {"dataset_value": value("train")},
"test_accuracy": {"dataset_value": value("test")},
}
)
def accuracy_computation(
dataset_value: str, embedded_data_set: pd.DataFrame
) -> tuple[float, float]:
"""Computed accuracy and standard error for a given dataset.
The accuracy I compute is the accuracy of a simple rule that predicts 'similar (1)' if the cosine similarity is
above some threshold X and otherwise predicts 'dissimilar (0)'.
:param dataset_value: train or test.
:param embedded_data_set: the embedded data set to compute accuracy on given the dataset_value..
:return: tuple of accuracy and standard error
"""
data = embedded_data_set[embedded_data_set["dataset"] == dataset_value]
a, se = _accuracy_and_se(data["cosine_similarity"], data["label"])
return a, se
def _embedding_multiplied_by_matrix(embedding: List[float], matrix: torch.tensor) -> np.array:
"""Helper function to multiply an embedding by a matrix."""
embedding_tensor = torch.tensor(embedding).float()
modified_embedding = embedding_tensor @ matrix
modified_embedding = modified_embedding.detach().numpy()
return modified_embedding
def _apply_matrix_to_embeddings_dataframe(matrix: torch.tensor, df: pd.DataFrame):
"""Helper function to compute new embeddings given a matrix, and then compute new cosine similarities.
Note: this mutates the passed in dataframe.
"""
# create custom embeddings
for column in ["text_1_embedding", "text_2_embedding"]:
df[f"{column}_custom"] = df[column].apply(
lambda x: _embedding_multiplied_by_matrix(x, matrix)
)
# compute cosine similarities
df["cosine_similarity_custom"] = df.apply(
lambda row: _cosine_similarity(
row["text_1_embedding_custom"], row["text_2_embedding_custom"]
),
axis=1,
)
optimization_parameterization = {
"matrix_b10_l10": {"batch_size": value(10), "learning_rate": value(10.0)},
"matrix_b100_l100": {"batch_size": value(100), "learning_rate": value(100.0)},
"matrix_b1000_l1000": {"batch_size": value(1000), "learning_rate": value(1000.0)},
}
@parameterize(**optimization_parameterization)
def optimize_matrix(
embedded_data_set: pd.DataFrame,
modified_embedding_length: int = 2048,
batch_size: int = 100,
max_epochs: int = 10, # set to this while initially exploring
learning_rate: float = 100.0, #
dropout_fraction: float = 0.0, #
print_progress: bool = True,
save_results: bool = True,
) -> pd.DataFrame:
"""Return matrix optimized to minimize loss on training data.
:param embedded_data_set: the embedding data set to optimize.
:param modified_embedding_length: length of new embedding. From the cookbook - bigger was better (2048 is length of babbage encoding).
:param batch_size: size of the batch to optimize.
:param max_epochs: max number of epochs. Set to this small while initially exploring
:param learning_rate: the learning rate to use. From the cookbook - seemed to work best when similar to batch size - feel free to try a range of values.
:param dropout_fraction: From the cookbook - dropout helped by a couple percentage points (definitely not necessary).
:param print_progress:
:param save_results:
:return: dataframe cataloging the optimization results.
"""
run_id = random.randint(0, 2**31 - 1) # (range is arbitrary)
# convert from dataframe to torch tensors
# e is for embedding, s for similarity label
def tensors_from_dataframe(
df: pd.DataFrame,
embedding_column_1: str,
embedding_column_2: str,
similarity_label_column: str,
) -> Tuple[torch.tensor, torch.tensor, torch.tensor]:
e1 = np.stack(np.array(df[embedding_column_1].values))
e2 = np.stack(np.array(df[embedding_column_2].values))
s = np.stack(np.array(df[similarity_label_column].astype("float").values))
e1 = torch.from_numpy(e1).float()
e2 = torch.from_numpy(e2).float()
s = torch.from_numpy(s).float()
return e1, e2, s
e1_train, e2_train, s_train = tensors_from_dataframe(
embedded_data_set[embedded_data_set["dataset"] == "train"],
"text_1_embedding",
"text_2_embedding",
"label",
)
e1_test, e2_test, s_test = tensors_from_dataframe(
embedded_data_set[embedded_data_set["dataset"] == "test"],
"text_1_embedding",
"text_2_embedding",
"label",
)
# create dataset and loader
dataset = torch.utils.data.TensorDataset(e1_train, e2_train, s_train)
train_loader = torch.utils.data.DataLoader(dataset, batch_size=batch_size, shuffle=True)
# define model (similarity of projected embeddings)
def model(embedding_1, embedding_2, matrix, dropout_fraction=dropout_fraction):
e1 = torch.nn.functional.dropout(embedding_1, p=dropout_fraction)
e2 = torch.nn.functional.dropout(embedding_2, p=dropout_fraction)
modified_embedding_1 = e1 @ matrix # @ is matrix multiplication
modified_embedding_2 = e2 @ matrix
similarity = torch.nn.functional.cosine_similarity(
modified_embedding_1, modified_embedding_2
)
return similarity
# define loss function to minimize
def mse_loss(predictions, targets):
difference = predictions - targets
return torch.sum(difference * difference) / difference.numel()
# initialize projection matrix
embedding_length = len(embedded_data_set["text_1_embedding"].values[0])
matrix = torch.randn(embedding_length, modified_embedding_length, requires_grad=True)
epochs, types, losses, accuracies, matrices = [], [], [], [], []
for epoch in range(1, 1 + max_epochs):
# iterate through training dataloader
for a, b, actual_similarity in train_loader:
# generate prediction
predicted_similarity = model(a, b, matrix)
# get loss and perform backpropagation
loss = mse_loss(predicted_similarity, actual_similarity)
loss.backward()
# update the weights
with torch.no_grad():
matrix -= matrix.grad * learning_rate
# set gradients to zero
matrix.grad.zero_()
# calculate test loss
test_predictions = model(e1_test, e2_test, matrix)
test_loss = mse_loss(test_predictions, s_test)
# compute custom embeddings and new cosine similarities
_apply_matrix_to_embeddings_dataframe(matrix, embedded_data_set)
# calculate test accuracy
for dataset in ["train", "test"]:
data = embedded_data_set[embedded_data_set["dataset"] == dataset]
a, se = _accuracy_and_se(data["cosine_similarity_custom"], data["label"])
# record results of each epoch
epochs.append(epoch)
types.append(dataset)
losses.append(loss.item() if dataset == "train" else test_loss.item())
accuracies.append(a)
matrices.append(matrix.detach().numpy())
# optionally print accuracies
if print_progress is True:
print(
f"Epoch {epoch}/{max_epochs}: {dataset} accuracy: {a:0.1%} ± {1.96 * se:0.1%}"
)
data = pd.DataFrame({"epoch": epochs, "type": types, "loss": losses, "accuracy": accuracies})
data["run_id"] = run_id
data["modified_embedding_length"] = modified_embedding_length
data["batch_size"] = batch_size
data["max_epochs"] = max_epochs
data["learning_rate"] = learning_rate
data["dropout_fraction"] = dropout_fraction
data["matrix"] = matrices # saving every single matrix can get big; feel free to delete/change
# TODO: pull this out
if save_results is True:
data.to_csv(f"{run_id}_optimization_results.csv", index=False)
return data
# this injects all the results of the parameterizations of the optimize_matrix function
@inject(
optimization_result_matrices=group(*[source(k) for k in optimization_parameterization.keys()])
)
def optimization_results(
optimization_result_matrices: List[pd.DataFrame],
) -> pd.DataFrame:
"""Combine optimization results into one dataframe."""
return pd.concat(optimization_result_matrices)
def training_and_test_loss_plot(optimization_results: pd.DataFrame) -> go.Figure:
"""Plot training and test loss over time."""
# plot training loss and test loss over time
_plot = px.line(
optimization_results,
line_group="run_id",
x="epoch",
y="loss",
color="type",
hover_data=["batch_size", "learning_rate", "dropout_fraction"],
facet_row="learning_rate",
facet_col="batch_size",
width=500,
)
return _plot
def accuracy_plot(optimization_results: pd.DataFrame) -> go.Figure:
"""Plot accuracy over time."""
# plot accuracy over time
_plot = px.line(
optimization_results,
line_group="run_id",
x="epoch",
y="accuracy",
color="type",
hover_data=["batch_size", "learning_rate", "dropout_fraction"],
facet_row="learning_rate",
facet_col="batch_size",
width=500,
)
return _plot
def best_matrix(optimization_results: pd.DataFrame) -> pd.DataFrame:
"""Return the best matrix from the optimization results."""
best_run = optimization_results.sort_values(by="accuracy", ascending=False).iloc[0]
best_matrix = best_run["matrix"]
return best_matrix
def customized_embeddings_dataframe(
embedded_data_set: pd.DataFrame, best_matrix: pd.DataFrame
) -> pd.DataFrame:
"""Apply the best matrix to the embedded data set and return a dataframe with customized embeddings."""
_apply_matrix_to_embeddings_dataframe(best_matrix, embedded_data_set)
return embedded_data_set
def customized_dataset_histogram(
customized_embeddings_dataframe: pd.DataFrame,
) -> go.Figure:
"""Plot histogram of cosine similarities for the new customized embeddings.
The graphs show how much the overlap there is between the distribution of cosine similarities for similar and
dissimilar pairs. If there is a high amount of overlap, that means there are some dissimilar pairs with greater
cosine similarity than some similar pairs.
"""
_plot = px.histogram(
customized_embeddings_dataframe,
x="cosine_similarity_custom",
color="label",
barmode="overlay",
width=500,
facet_row="dataset",
)
return _plot
def test_accuracy_post_optimization(
customized_embeddings_dataframe: pd.DataFrame,
) -> tuple:
data = customized_embeddings_dataframe[customized_embeddings_dataframe["dataset"] == "test"]
a, se = _accuracy_and_se(data["cosine_similarity_custom"], data["label"])
print(f"test accuracy after optimization: {a:0.1%} ± {1.96 * se:0.1%}")
return a, se
if __name__ == "__main__":
import __main__ as customize_embeddings
logging.basicConfig(level=logging.INFO, stream=sys.stdout)
from hamilton import driver
dr = driver.Builder().with_modules(customize_embeddings).with_config({"source": "snli"}).build()
dr.display_all_functions(
"dag", render_kwargs={"format": "png"}, orient="TB", deduplicate_inputs=True
)
result = dr.execute(
[
"train_accuracy",
"test_accuracy",
"embedded_dataset_histogram",
"test_accuracy_post_optimization",
"accuracy_plot",
"training_and_test_loss_plot",
"customized_embeddings_dataframe",
"customized_dataset_histogram",
],
inputs={},
)
print(result["train_accuracy"])
print(result["test_accuracy"])
print(result["train_accuracy"])
print(result["test_accuracy_post_optimization"])
Requirements
numpy
openai
pandas
pandera
plotly
requests
scikit-learn
sf-hamilton
tenacity
torch